class: center, middle, inverse, title-slide # R Markdown Workshop ## Creat Reports with R Markdown ### Mayan Navon & Michael Pinus ### 18/07/2018 --- ## R Markdown - What is R Markdown - Possible outputs - PDF - Docx - HTML pages - Complete websites - Slideshows (HTML, or PP with RStudio>1.2) - Many more.. --- ## Required/Reviewed Software - MiKTeX (complete version - takes a long time to install) - RStudio + R - Output Viewer (Powerpoint, Word, Browser...) - R packages - CRAN: rmarkdown, knitr, kableExtra, ggpubr, GGally, rmdformats, rticles,citr (optional), pacman and installr (very optional, but useful) - Github: crsh/papaja, pinusm/Mmisc (optional) - Zotero (optional) --- ## Installing - Copy&Paste to R: - Might take a while... ```r install.packages("pacman") ``` ```r pacman::p_load(tidyverse,rmarkdown,knitr,citr,installr, kableExtra,ggpubr,GGally,rmdformats, rticles) pacman::p_load_gh("crsh/papaja","pinusm/Mmisc") ``` ```r installr::install.MikTeX() ``` --- ## Basic Workflow - Create new R Markdown file - Edit header (and other meta-data) - Analysis & Write Body - Knit - Revise (IN R MARKDOWN, not output) - Final adjustment on output (e.g. fix table font size with Word Macro) --- ## Meta-Data (YAML) - Basic formats ```r --- title: "My document" output: html_document pdf_document --- ``` - *rmdformats* HTML document ```r --- title: "atgn4 - summary report" author: "Mayan Navon" date: 'June 20th, 2018' # '`r format(Sys.Date(), "%d/%m/%Y")`' output: rmdformats::readthedown: highlight: kate fig_height: 4 fig_width: 5 html_document: df_print: paged theme: cerulean --- ``` --- ## Analysis & Write Body - Write body - Write in plain text - Use markdown syntax to format - Insert chunks (per analysis/ data preparation/ other data handling) - insert: R - Edit chunk parameters - **Code evaluation:** eval, include - **Results:** echo, message, warning - Many more... see reference guide --- ## Tables knitr::kable() and kableExtra::kable_styling() ```r '''{r demo table, echo = T, warning=FALSE,message=F} pacman::p_load(afex) # for demo data and analysis data(sk2011.1) dt <- aov_ez("id", "response", sk2011.1, between = "instruction", within = "plausibility", anova_table = list(correction = "none", es = "pes") ) ''' ``` ''' should be BACKTICKS!! --- ## knitr::kable() ```r kable(afex::nice(dt)) ``` <table> <thead> <tr> <th style="text-align:left;"> Effect </th> <th style="text-align:left;"> df </th> <th style="text-align:left;"> MSE </th> <th style="text-align:left;"> F </th> <th style="text-align:left;"> pes </th> <th style="text-align:left;"> p.value </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> instruction </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 506.86 </td> <td style="text-align:left;"> 0.31 </td> <td style="text-align:left;"> .008 </td> <td style="text-align:left;"> .58 </td> </tr> <tr> <td style="text-align:left;"> plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 34.23 *** </td> <td style="text-align:left;"> .47 </td> <td style="text-align:left;"> <.0001 </td> </tr> <tr> <td style="text-align:left;"> instruction:plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 10.67 ** </td> <td style="text-align:left;"> .22 </td> <td style="text-align:left;"> .002 </td> </tr> </tbody> </table> --- ## kableExtra::kable_styling() ```r kable(afex::nice(dt)) %>% kable_styling( bootstrap_options = c("striped", "hover", "condensed", "responsive"), #bootstrap_options ignored in Slides output, but are useful full_width = F) ``` <table class="table table-striped table-hover table-condensed table-responsive" style="width: auto !important; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Effect </th> <th style="text-align:left;"> df </th> <th style="text-align:left;"> MSE </th> <th style="text-align:left;"> F </th> <th style="text-align:left;"> pes </th> <th style="text-align:left;"> p.value </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> instruction </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 506.86 </td> <td style="text-align:left;"> 0.31 </td> <td style="text-align:left;"> .008 </td> <td style="text-align:left;"> .58 </td> </tr> <tr> <td style="text-align:left;"> plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 34.23 *** </td> <td style="text-align:left;"> .47 </td> <td style="text-align:left;"> <.0001 </td> </tr> <tr> <td style="text-align:left;"> instruction:plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 10.67 ** </td> <td style="text-align:left;"> .22 </td> <td style="text-align:left;"> .002 </td> </tr> </tbody> </table> --- ## kableExtra::add_header_above() ```r kable(afex::nice(dt)) %>% kable_styling( bootstrap_options = c("striped", "hover", "condensed", "responsive"), full_width = F) %>% add_header_above(c("Header Spanning 3 cols" = 3, " " = 3)) %>% add_header_above(c("Header Spanning 6 cols" = 6)) ``` <table class="table table-striped table-hover table-condensed table-responsive" style="width: auto !important; margin-left: auto; margin-right: auto;"> <thead> <tr><th style="border-bottom:hidden; padding-bottom:0; padding-left:3px;padding-right:3px;text-align: center; " colspan="6"><div style="border-bottom: 1px solid #ddd; padding-bottom: 5px;">Header Spanning 6 cols</div></th></tr> <tr> <th style="border-bottom:hidden; padding-bottom:0; padding-left:3px;padding-right:3px;text-align: center; " colspan="3"><div style="border-bottom: 1px solid #ddd; padding-bottom: 5px;">Header Spanning 3 cols</div></th> <th style="border-bottom:hidden" colspan="3"></th> </tr> <tr> <th style="text-align:left;"> Effect </th> <th style="text-align:left;"> df </th> <th style="text-align:left;"> MSE </th> <th style="text-align:left;"> F </th> <th style="text-align:left;"> pes </th> <th style="text-align:left;"> p.value </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> instruction </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 506.86 </td> <td style="text-align:left;"> 0.31 </td> <td style="text-align:left;"> .008 </td> <td style="text-align:left;"> .58 </td> </tr> <tr> <td style="text-align:left;"> plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 34.23 *** </td> <td style="text-align:left;"> .47 </td> <td style="text-align:left;"> <.0001 </td> </tr> <tr> <td style="text-align:left;"> instruction:plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 10.67 ** </td> <td style="text-align:left;"> .22 </td> <td style="text-align:left;"> .002 </td> </tr> </tbody> </table> --- ## kableExtra::footnote() ```r kable(afex::nice(dt)) %>% kable_styling( bootstrap_options = c("striped", "hover", "condensed", "responsive"), full_width = F) %>% footnote(general = " Footnote A", footnote_as_chunk = T) ``` <table class="table table-striped table-hover table-condensed table-responsive" style="width: auto !important; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Effect </th> <th style="text-align:left;"> df </th> <th style="text-align:left;"> MSE </th> <th style="text-align:left;"> F </th> <th style="text-align:left;"> pes </th> <th style="text-align:left;"> p.value </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> instruction </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 506.86 </td> <td style="text-align:left;"> 0.31 </td> <td style="text-align:left;"> .008 </td> <td style="text-align:left;"> .58 </td> </tr> <tr> <td style="text-align:left;"> plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 34.23 *** </td> <td style="text-align:left;"> .47 </td> <td style="text-align:left;"> <.0001 </td> </tr> <tr> <td style="text-align:left;"> instruction:plausibility </td> <td style="text-align:left;"> 1, 38 </td> <td style="text-align:left;"> 117.21 </td> <td style="text-align:left;"> 10.67 ** </td> <td style="text-align:left;"> .22 </td> <td style="text-align:left;"> .002 </td> </tr> </tbody> <tfoot><tr><td style="padding: 0; border: 0;" colspan="100%"> <span style="font-style: italic;">Note: </span> <sup></sup> Footnote A</td></tr></tfoot> </table> --- ## Figures - ggpubr:: - GGally:: ```r data(flea) df <- flea df %>% head(5) ``` ``` ## species tars1 tars2 head aede1 aede2 aede3 ## 1 Concinna 191 131 53 150 15 104 ## 2 Concinna 185 134 50 147 13 105 ## 3 Concinna 200 137 52 144 14 102 ## 4 Concinna 173 127 50 144 16 97 ## 5 Concinna 171 118 49 153 13 106 ``` ```r df %>% select(species) %>% table() ``` ``` ## . ## Concinna Heikert. Heptapot. ## 21 31 22 ``` --- ## Some fixes before plotting ```r df <- flea %>% rename(`Head Diameter` = head, `1st Torso Diameter` = tars1, `2nd Torso Diameter` = tars2, Species = species) %>% mutate(Species = fct_recode(Species, "Heikert" = "Heikert.", "Heptapot" = "Heptapot.") ) ``` --- ## ggpubr::violin() ```r my_comparisons <- list( c("Heptapot", "Heikert"), c("Heikert", "Concinna"), c("Heptapot", "Concinna")) ``` .pull-left[ 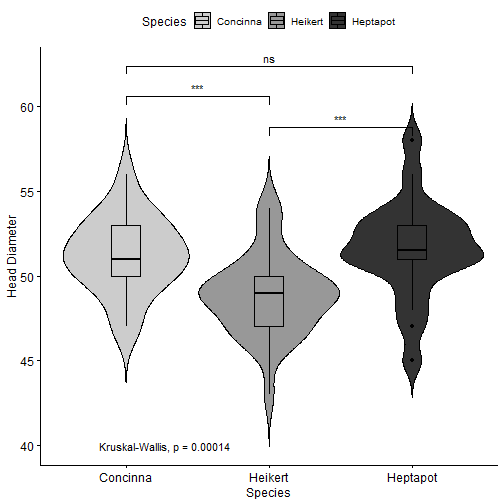 ] .pull-right[ ```r ggviolin(df, x = "Species", y = "`Head Diameter`", fill = "Species", palette = "grey", add = "boxplot", dd.params = list( fill = "white") ) + # Add significance levels stat_compare_means( comparisons = my_comparisons, label = "p.signif" ) + # Add the global p-value stat_compare_means( label.y = 40 ) ``` ] --- ## ggpubr::ggerrorplot() ```r ggerrorplot(df, x = "Species", y = "`Head Diameter`", color = "Species", desc_stat = "mean_ci", palette= "grey",position = position_dodge(0.3), size = 0.9) ``` 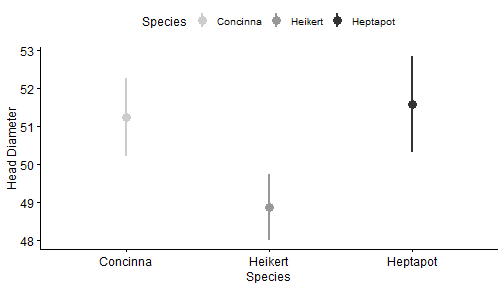 --- ## GGally::ggpairs() ```r ggpairs(df, columns = 2:4, lower = list(continuous = Mmisc::plot_trend_lines)) + papaja::theme_apa() ``` 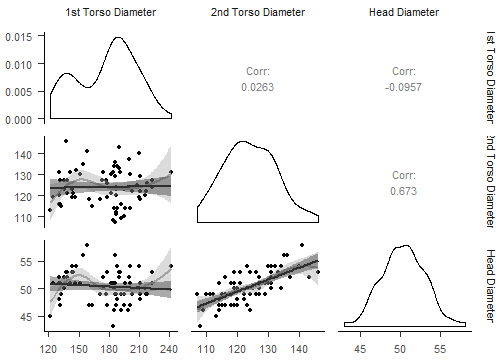 --- ## knitr::include_graphics() When plots are not generated from R code, we generate the images manually and pass their file paths to `include_graphics()` to include them in the output. ```r knitr::include_graphics("figures/poweranalysis.png") ``` <img src="figures/poweranalysis.png" width="522" /> --- ## Latex We can use `\(\LaTeX\)` tricks! Possible examples include: ```r This sentence contains normal and $\textit{italic text}$. ``` This sentence contains normal and `\(\textit{italic text}\)`. <br><br><br> and also a bit more complex stuff: ```r $$\begin{align} y|x &\sim Poisson(\lambda(x)) \end{align}$$ ``` `$$\begin{align} y|x &\sim Poisson(\lambda(x)) \end{align}$$` --- ## Inline Text ```r dt_print <- apa_print(dt , es = "pes") ``` We might want to reference some data in our text. We _could_ type it.. (Note the use of `\(\LaTeX\)`) ```r The effect of Instruction on response was not significant *F*(1, 38) = 0.31, $\mathit{MSE}$ = 506.86, *p* = .583, $\hat{\eta}^2_p = .008 ``` prints out as: The effect of Instruction on response was not significant `\(F(1, 38) = 0.31\)`, `\(\mathit{MSE} = 506.86\)`, `\(p = .583\)`, `\(\hat{\eta}^2_p = .008\)` --- ## Inline Text - Cont. We can also automagically insert it: ```r The effect of Instruction on response was not significant `r dt_print$full_result$instruction` ``` prints out as: The effect of Instruction on response was not significant `\(F(1, 38) = 0.31\)`, `\(\mathit{MSE} = 506.86\)`, `\(p = .583\)`, `\(\hat{\eta}^2_p = .008\)` --- ## Some output formats .pull-left[ #### rmdformats::htmlclean <img src="figures/rmdformats_htmlclean.jpg" width="1707" /> #### rmdformats::readthedown <img src="figures/rmdformats_readthedown.jpg" width="1707" /> ] .pull-right[ #### rticles::plos <img src="figures/plos.png" width="1555" /> #### rticles::PNAS <img src="figures/PNAS.png" width="1495" /> ] --- ## Knit - Click the *Knit* button at the top of the RStudio scripts pane, OR: ```r rmarkdown::render("<file path>") ``` --- ## papaja Tables ### papaja::apa_print() and papaja::apa_table() ```r dt_print <- apa_print(dt , es = "pes") apa_table(dt_print$table) ``` A bit of cheating: - This is a screenshoted Docx document - We ran a Macro to fix font size and line spacing (we'll cover that later) <img src="figures/papaja_cheat.png" width="1837" /> --- ## papaja::apa_table() with caption and note ```r apa_table(dt_print$table , note = "this is a note", caption = "this is a caption") ``` Same cheats as before... <img src="figures/papaja_cheat_footnote.png" width="1843" /> --- ## Figure Captions and Cross-References <img src="figures/papaja_cheat_plots.png" width="474" /> ```r In papaja we can also create cross-references, such as this. See Figure \@ref(fig:includegraphics). ``` --- ## Citations We can cite bibliography records from `Zotero` using the `citr` addin in rstudio. <img src="figures/citr.png" width="468" /> --- ## Papaja Workflow Useful when the analysis takes more than a few seconds to run. - Analysis as usual (R) - Get numbers (*M*s/*SD*s/etc), tables and figures to (at least almost) console-priniting shape - Save R environment to a file (e.g., "analysis.Rdata") - Create new R Markdown file - Edit header - Load R environment from file (e.g., "analysis.Rdata") - Write Body (+ cite previous works) - Knit - Revise (IN R MARKDOWN, not output) - Final adjustment on output (e.g. fix table font size with Word Macro) --- ## Fix tables with Word Macro ```VBA Sub fixPapajaTables() ' ' fixPapajaTables Macro ' ' Dim tbl As Table For Each tbl In ActiveDocument.Tables tbl.Range.Font.Size = 10 tbl.Range.ParagraphFormat.LineSpacing = 10 Next End Sub ``` ## Non-R Workflow - Analysis as usual - Get numbers (*M*s/*SD*s/etc) - Save tables and figures as PNG/JPG graphic files - Create new R Markdown file - ... - Write Body (insert numbers and graphics) - ... - Final adjustment on output (probably not needed as the graphics were embedded as-is) Remember non-R elements can be combined in an R workflow